Structural Basis for Double-Stranded RNA Processing by Dicer
RNA interference is one of the hotspots in biological studies these days. People keep trying to apply it as a new research tool and a new drug of defeating diseases. However, its molecular mechanism has still not been fully understood, which prevents its whole practical potential from being developed. This article reveals the crystal structure of dicer from Giardia intestinalis; makes us one step further to fully understanding of the initiation step of RNAi.
Firstly, the authors decided to use dicer from Giardia intestinalis, a flagellated protozoan parasite, rather than human dicer. Since the former one is smaller and have the intact enzyme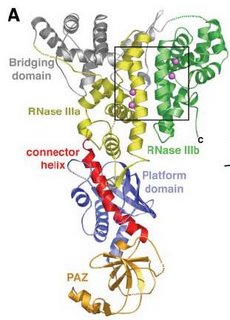 activity, the whole work can be much simpler. The Giardia Dicer has six domains (as labeled in the right figure) and looks like a hatchet from this direction. The two RNase III domains form the blade, the PAZ domain makes up the base of the handle and the connector helix connects the two parts, while platform domain wraps the connector helix.
activity, the whole work can be much simpler. The Giardia Dicer has six domains (as labeled in the right figure) and looks like a hatchet from this direction. The two RNase III domains form the blade, the PAZ domain makes up the base of the handle and the connector helix connects the two parts, while platform domain wraps the connector helix.
According to the authors' study, the two RNase III domains have two metal binding sites each (purple spheres in the box), which are considered as two-metal-ion cleavage sites for dsRNA. PAZ domain can bind the end of dsRNA with its 3' two-nucleotide RNA binding pocket. The distance from two-metal-ion cleavage sites of RNase III to the two-nucleotide RNA binding pocket of PAZ domain is ~65 Å, which matches the length of 25 dsRNA base pairs. Here the connector helix seems to play an important role in determining the length of siRNA to be cleaved. Variant lengths of connector helix thus cause different lengths of siRNA being produced.
With this article, several gaps of understanding of how dicer works in RNAi were filled, several previous observations were able to be explained.
Wanna know more about it? Wait for my literature review! It's com ing soon!
ing soon!
Chen
Firstly, the authors decided to use dicer from Giardia intestinalis, a flagellated protozoan parasite, rather than human dicer. Since the former one is smaller and have the intact enzyme
 activity, the whole work can be much simpler. The Giardia Dicer has six domains (as labeled in the right figure) and looks like a hatchet from this direction. The two RNase III domains form the blade, the PAZ domain makes up the base of the handle and the connector helix connects the two parts, while platform domain wraps the connector helix.
activity, the whole work can be much simpler. The Giardia Dicer has six domains (as labeled in the right figure) and looks like a hatchet from this direction. The two RNase III domains form the blade, the PAZ domain makes up the base of the handle and the connector helix connects the two parts, while platform domain wraps the connector helix.According to the authors' study, the two RNase III domains have two metal binding sites each (purple spheres in the box), which are considered as two-metal-ion cleavage sites for dsRNA. PAZ domain can bind the end of dsRNA with its 3' two-nucleotide RNA binding pocket. The distance from two-metal-ion cleavage sites of RNase III to the two-nucleotide RNA binding pocket of PAZ domain is ~65 Å, which matches the length of 25 dsRNA base pairs. Here the connector helix seems to play an important role in determining the length of siRNA to be cleaved. Variant lengths of connector helix thus cause different lengths of siRNA being produced.
With this article, several gaps of understanding of how dicer works in RNAi were filled, several previous observations were able to be explained.
Wanna know more about it? Wait for my literature review! It's com
 ing soon!
ing soon!Chen

0 Comments:
Post a Comment
<< Home